Equivalence with bacterial reference strains
Thousands of living species are newly described every year and in the microbial world the tree of life is expanding dramatically due to the identification of previously unknown lineages through genome analysis 1. For bacteria, the first isolate of a confirmed new species, the type strain, must be deposited within two internationally recognised culture collections, i.e. collections such as NCTC that meet the OECD Best Practice Guidelines for Biological Resource Centres.
Campylobacter hepaticus, a new species isolated recently in Australia from the livers of chickens with spotty liver disease4, was deposited with NCTC, as NCTC 13823, and also with the Collection de l’Institut Pasteur (CIP), as CIP 111092. The original depositor’s identifier for the strain, HV10, is cross-referenced by both collections. NCTC 13823 and CIP 111092 can therefore be described as equivalent reference strains.
Historically, reference cultures were circulated between collections to make the strains as widely accessible as possible for the scientific community. This means that a single strain can have multiple equivalents. NCTC 9001, the type strain of Escherichia coli, has equivalents in culture collections across Europe, Japan and the US. Demonstration of equivalence is important not only using classical tests5, but also using emerging technologies including whole genome sequencing and bioinformatic techniques. Internationally recognised methods often specify use of strains from a single culture collection without cross-referencing the equivalent reference strains, which means scientists may be discouraged from using their local or most accessible collection.
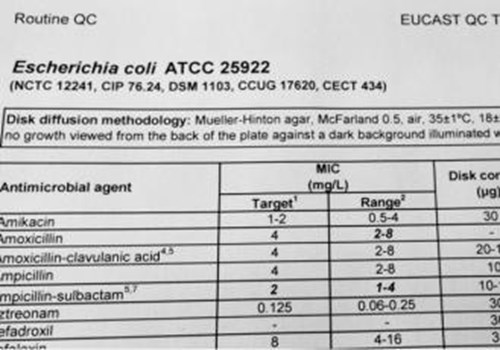
The European Committee on Antimicrobial Susceptibility Testing (EUCAST) stipulates the control strains to be used for MIC determination and disk diffusion and although the strain numbers from the US collection, ATCC, have more prominence, the equivalent strains in recognised European collections, including NCTC, are also listed.
By contrast, the three compendia, the United States, European, and Japanese Pharmacopoeia, used widely for pharmaceutical quality control, tend to stipulate ATCC numbers only. It is less clearly denoted that pharmacopoeia users may adopt alternative methods with agreement from their competent authority. For example, in the UK this could be a licensing authority such as the Medicines & Healthcare products Regulatory Agency (MHRA).
The technical committee that developed ISO standard 11133:2014 (Microbiology of food, animal feed and water – preparation, production, storage and performance testing of culture media) has gone further in addressing equivalence between culture collections by referring to the Global Catalogue of Microorganisms which lists a wide range of equivalent strains using World Data Centre for Microorganisms (WDCM) identifiers. However, it must be noted that not every collection listed in the Global Catalogue meets the OECD Best Practice Guidelines.
The identification numbers for NCTC strain equivalents in other collections are provided in the NCTC electronic catalogue. Another way of finding which NCTC strain is equivalent to an ATCC control microorganism listed in a standard, an ISO method or a pharmacopoeia, is to search by bacterial strain and cross referencing the ATCC strain number via the link below:
NCTC equivalents to ATCC bacterial strains
References
- Laura A. Hug et al., 2016. A new view of the tree of life. Nature Microbiology, 1(5), p.16048. http://www.nature.com/articles/nmicrobiol201648.pdf
- Van, T.T.H. et al., 2016. Campylobacter hepaticus sp. nov., isolated from chickens with spotty liver disease. International journal of systematic and evolutionary microbiology, 66(11), pp.4518–4524. http://ijs.microbiologyresearch.org/content/journal/ijsem/10.1099/ijsem.0.001383
- Cundell A. Chatellier et al., 2010. Equivalence of quality control strains of microorganisms used in the compendial microbiological tests: are national culture collection strains identical? PDA J Pharm Sci Technol, 64(2), p137-55.
https://www.ncbi.nlm.nih.gov/pubmed/21502014
